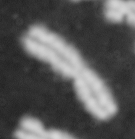
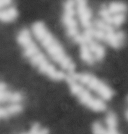
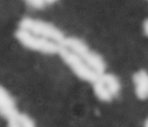
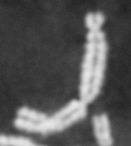
Please note: This webpage is intended for analysis of individual chromosomes and has been superceded by the full ADCI software. ADCI automatically selects metaphase images and determines radiation dose. To read more about ADCI or to obtain a demonstration version of the software, visit radiation.cytognomix.com.
Automated Dicentric Chromosome Identification
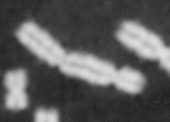
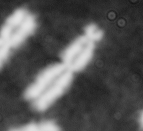
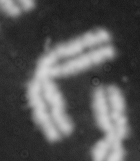
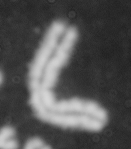
Dose from radiation exposure can be estimated from dicentric chromosome (DC) frequencies in metaphase cells of peripheral blood lymphocytes. We automated DC detection by extracting features in Giemsa-stained metaphase chromosome images and classifying objects by machine learning (ML). DC detection involves (i) intensity thresholded segmentation of metaphase objects, (ii) chromosome separation by watershed transformation and elimination of inseparable chromosome clusters, fragments and staining debris using a morphological decision tree filter, (iii) determination of chromosome width and centreline, (iv) derivation of centromere candidates, and (v) distinction of DCs from monocentric chromosomes (MC) by ML. Centromere candidates are inferred from 14 image features input to a Support Vector Machine (SVM). Sixteen features derived from these candidates are then supplied to a Boosting classifier and a second SVM which determines whether a chromosome is either a DC or MC. The SVM was trained with 292 DCs and 3135 MCs, and then tested with cells exposed to either low (1 Gy) or high (2-4 Gy) radiation dose. Results were then compared with those of 3 experts. True positive rates (TPR) and positive predictive values (PPV) were determined for the tuning parameter, σ. At larger σ, PPV decreases and TPR increases. At high dose, for σ = 1.3, TPR = 0.52 and PPV = 0.83, while at σ = 1.6, the TPR = 0.65 and PPV = 0.72. At low dose and σ = 1.3, TPR = 0.67 and PPV = 0.26. The algorithm differentiates DCs from MCs, overlapped chromosomes and other objects with acceptable accuracy over a wide range of radiation exposures.
Author Information
Yanxin LI1, Joan H. Knoll2,3, Ruth C. Wilkins4, Farrah N. Flegal5 and Peter K. Rogan1,3,*
1. Department of Biochemistry, Schulich School of Medicine and Dentistry, University of Western Ontario, London, Ontario N6A 5C1, Canada2. Department of Pathology and Laboratory Medicine, Schulich School of Medicine and Dentistry, University of Western Ontario
3. Cytognomix Inc, London, Ontario N5X 3X5, Canada
4. Consumer and Clinical Radiation Protection Bureau, Health Canada, Ottawa, Ontario, Canada
5. Biodosimetry Emergency Response Laboratory, Canadian Nuclear Laboratories
*Correspondence to: Peter K. Rogan, Department of Biochemistry, University of Western Ontario, DSB 5012, Schulich School of Medicine and Dentistry, London, Ontario N6A 5C1 Canada. E-mail: progan@uwo.ca
Try the Dicentric Chromosome Identifier web app
Image upload has been discontinued and has been superceded by the full ADCI software. The "Chromosome samples gallery" section below can still be utilized to view algorithm results. A demonstration version of ADCI can be downloaded from radiation.cytognomix.com.